Interpretable Word Embeddings from knowledge graph embeddings
A while ago, I created interpretable word embeddings using polar opposites (I used their jupyter notebook from here https://github.com/Sandipan99/POLAR) from wikidata5m knowledge graph embeddings (from here: https://graphvite.io/docs/latest/pretrained_model.html). It resulted in a gigantic file of pretrained embeddings which sort concepts along 700 semantic differentials, i.e. like good/bad.
However, the wikidata5m knowledge graph is huge. Roundabout 5 million concepts and 13 million spellings. A joined parquet file would properly take 100 GB of disk space. One way to make it usable with limited RAM is using databases. I could just use duckdb or arrow and tidytext together. I don’t like the thought of using a huge file just to assign embeddings. Also lot’s of the concepts in the knowledge graph are exotic, the tokens rarely attract human interest. Like specific fighter jet model spellings.
The appeal of this large graph is that it allows to create interpretable document embeddings and investigate corpora without the need for domain specific topic modeling, more concepts means more coverage.
All I want is that it is flexible enough to be used on real sentences, we can achieve that with a huge vocabulary or character n-grams.
I need to reduce the size to make it practical.
I had got inspired by radix.ai to reproject quantized fasttext embeddings into knowledge graph embeddings with an MLP before, this here seems like a persuasively valuable use case of that approach to try that out.
First step is to make the spaces mappable. For that we gotta create a subset of the knowledge graph embeddings which overlap with the learned terms from the fasttext embeddings (we can also expand that for select concepts). Let’s do that.
Taking a peek at the embeddings
To exemplify, I’ll grab an example concept and show how it is positioned.
pacman::p_load(tidytable, data.table, tidyverse, arrow, ggcharts, wordVectors, pbmcapply)
#Some file preparations
polar_embeddings <- arrow::read_parquet("/run/media/knut/HD/knut/Desktop/MLearningAlgoTests/data/polar/polar.parquet")
polar_ids <- fread("/run/media/knut/HD/MLearningAlgoTests/data/polar/polar_id.csv")
polar_embeddings$rowid <- polar_ids$id
entities <- fread("/run/media/knut/HD/MLearningAlgoTests/data/polar/entities.csv") %>% separate.(col = label, c("label2", "specification"), sep = "(", remove = F) %>% mutate.(label2=str_squish(label2))
entities %>% mutate(label2=tolower(label2)) %>% filter.(str_detect(label2, "sadam hussain"))## # A tidytable: 1 × 5
## rowid wikientity label label2 specification
## <dbl> <chr> <chr> <chr> <chr>
## 1 25635 Q1316 sadam hussain sadam hussain <NA>So how is this guy positioned?
On the polar opposites scale, the numbers can be negative.
Note this system is not perfect, not all scores make sense.
It can be a good starting point for forming content hypotheses.
subject <- polar_embeddings %>% filter.(rowid==entities %>% mutate(label2=tolower(label2)) %>% filter.(str_detect(label2, "sadam hussain")) %>% pull(rowid))
positioning_pos <- subject %>% t() %>% as.data.frame() %>% arrange(desc(V1))
positioning_pos$variable <- rownames(positioning_pos)
positioning_neg <- subject %>% t() %>% as.data.frame() %>% arrange(V1)
positioning_neg$variable <- rownames(positioning_neg)
show_me <- positioning_pos[1:20,] %>% bind_rows.(positioning_neg[1:20,]) %>% filter.(variable%in%c("sheep_wolf", "girl_men", "democratic_despotic", "pass_play", "confusion_peace", "bullet_safe"))
ggcharts::lollipop_chart(show_me, x = variable, y = V1)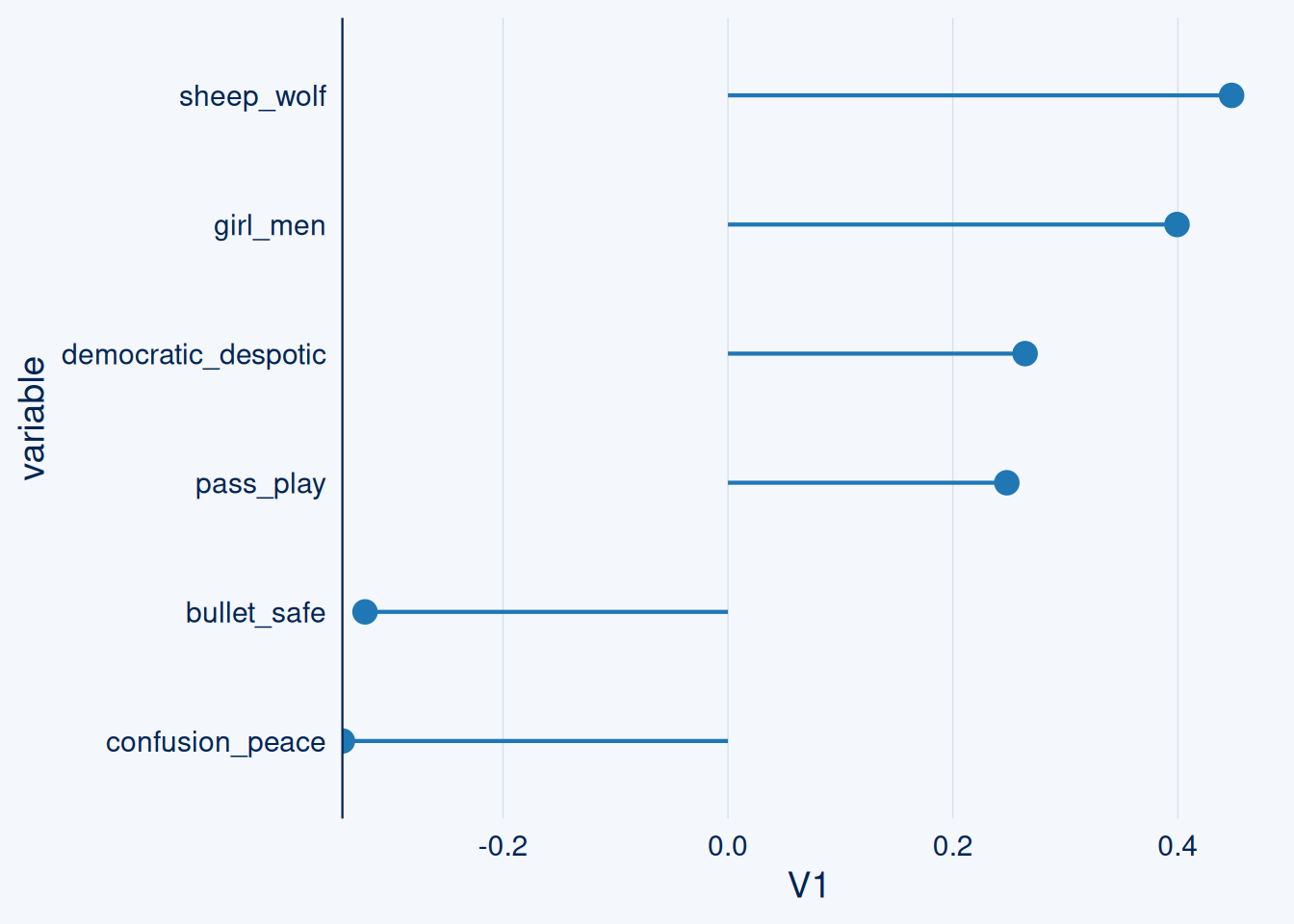
Getting the subset of directly mappable concepts
I got some quantized fasttext embeddings from here (https://zenodo.org/record/3629537). They’re just 1.2 gb, 2 million words.
It is clear we need automated disambiguation:
library(fastrtext)
model <- fastrtext::load_model("/home/knut/nlpaug/embs/cc.en.300.ftz")
words <- data.table(words=get_dictionary(model)) %>% filter(words%in%quanteda::stopwords(source = "smart")==F) %>% filter(nchar(words)>3)
entity_overlap <- entities %>% mutate.(words=tolower(label2)) %>%inner_join.(words)
entity_overlap %>% head()## # A tidytable: 6 × 6
## rowid wikientity label label2 specification words
## <dbl> <chr> <chr> <chr> <chr> <chr>
## 1 38101 Q3283450 time (third day album) time third day album) time
## 2 68619 Q385768 time (lionel richie album) time lionel richie album) time
## 3 74506 Q493981 time time <NA> time
## 4 323363 Q617572 time (yugoslavian band) time yugoslavian band) time
## 5 323363 Q617572 time (rock band) time rock band) time
## 6 323363 Q617572 time (yugoslav band) time yugoslav band) timeWe also have the descriptions of those entities from wikipedia. We can use the pretrained embeddings to disambiguate among those, which we’ll use as a quick and dirty way to disambiguate.
library(readr)
wikidata5m_text <- read_delim("/run/media/knut/HD/knut/Desktop/MLearningAlgoTests/data/wikidata5m_text.csv",
delim = "\t", escape_double = FALSE,
trim_ws = TRUE, col_names = FALSE) %>% rename(wikientity=X1, description=X2) %>% filter.(wikientity%in%unique(entity_overlap$wikientity))## Warning: One or more parsing issues, call `problems()` on your data frame for details,
## e.g.:
## dat <- vroom(...)
## problems(dat)## Rows: 4403676 Columns: 2
## ── Column specification ────────────────────────────────────────────────────────
## Delimiter: "\t"
## chr (2): X1, X2
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.wikidata5m_text %>% head()## # A tidytable: 6 × 2
## wikientity description
## <chr> <chr>
## 1 Q5596987 "Grape-Nuts is a breakfast cereal developed in 1897 by C. W. Post,…
## 2 Q3764405 "'Kadapa Lok Sabha constituency is one of the twenty-five lok sabh…
## 3 Q3991654 "TinyPic is a photo and video sharing service, owned and operated …
## 4 Q3990637 "Thomas Hedley Jr., (born 1942/43) is a British magazine editor an…
## 5 Q4929227 "Blue Hill Avenue is a regional rail station on the MBTA Commuter …
## 6 Q2291075 "Arabi (Arabic: أرابي) is a town in Crisp County, Georgia, United …Let’s test disambiguation via the fasttext model document embeddings:
test_entities <- entity_overlap %>% filter.(words=="communication")
entity_candidate_descr_embs <- get_sentence_representation(model, wikidata5m_text %>% filter.(wikientity%in%test_entities$wikientity) %>% pull(description))
entity_candidate_descr_embs <- entity_candidate_descr_embs %>% as.VectorSpaceModel()
rownames(entity_candidate_descr_embs) <- wikidata5m_text %>% filter.(wikientity%in%test_entities$wikientity) %>% distinct(wikientity) %>% pull(wikientity)
overlapping_words <- unique(entity_overlap$words)
fasttext_emb_test <- fastrtext::get_word_vectors(model, words = "communication")
fasttext_emb_test <- fasttext_emb_test %>% as.VectorSpaceModel()
rownames(fasttext_emb_test) <- "communication"
wordVectors::closest_to(entity_candidate_descr_embs, fasttext_emb_test) %>% rename(wikientity=word) %>% inner_join.(entity_overlap %>% distinct(wikientity, words)) %>% arrange(desc(`similarity to fasttext_emb_test`))## # A tidytable: 14 × 3
## wikientity `similarity to fasttext_emb_test` words
## <chr> <dbl> <chr>
## 1 Q11024 0.429 communication
## 2 Q11024 0.429 communicate
## 3 Q11024 0.429 communicating
## 4 Q11024 0.429 miscommunication
## 5 Q11024 0.429 comunication
## 6 Q18148644 0.394 communication
## 7 Q5154077 0.341 communication
## 8 Q1121530 0.335 communication
## 9 Q5154076 0.260 communication
## 10 Q5154087 0.253 communication
## 11 Q5154086 0.251 communication
## 12 Q333886 0.248 communication
## 13 Q7733071 0.219 communication
## 14 Q21161211 0.200 communicationThe description of the most similar concept is:
wikidata5m_text %>% filter.(wikientity=="Q11024") %>% pull(description)## [1] "Communication (from Latin communicare, meaning \"to share\") is the act of conveying meanings from one entity or group to another through the use of mutually understood signs, symbols, and semiotic rules.The main steps inherent to all communication are:The formation of communicative motivation or reason.Message composition (further internal or technical elaboration on what exactly to express).Message encoding (for example, into digital data, written text, speech, pictures, gestures and so on).Transmission of the encoded message as a sequence of signals using a specific channel or medium.Noise sources such as natural forces and in some cases human activity (both intentional and accidental) begin influencing the quality of signals propagating from the sender to one or more receivers.Reception of signals and reassembling of the encoded message from a sequence of received signals.Decoding of the reassembled encoded message.Interpretation and making sense of the presumed original message.The scientific study of communication can be divided into:Information theory which studies the quantification, storage, and communication of information in general;Communication studies which concerns human communication;Biosemiotics which examines communication in and between living organisms in general.The channel of communication can be visual, auditory, tactile (such as in Braille) and haptic, olfactory, electromagnetic, or biochemical.Human communication is unique for its extensive use of abstract language. Development of civilization has been closely linked with progress in telecommunication."In this instance, this simple approach worked. Let’s proceed with it.
# overlapping_words <- unique(entity_overlap$words)
#
#
# rm(polar_embeddings, entities)
# gc()
#
# pacman::p_load(tidytable, data.table, tidyverse, arrow, ggcharts, wordVectors, pbmcapply, fastrtext)
#
# fastSave::load.lbzip2("/home/knut/Desktop/image.RDataFS", n.cores = 15)
#
# disambiguate <- function(X){
# word <- X
# model <- fastrtext::load_model("/home/knut/nlpaug/embs/cc.en.300.ftz")
# test_entities <- entity_overlap %>% filter.(words==word)
#
# entity_candidate_descr_embs <- fastrtext::get_sentence_representation(model, wikidata5m_text %>% filter.(wikientity%in%test_entities$wikientity) %>% pull(description))
# entity_candidate_descr_embs <- entity_candidate_descr_embs %>% as.VectorSpaceModel()
# rownames(entity_candidate_descr_embs) <- wikidata5m_text %>% filter.(wikientity%in%test_entities$wikientity) %>% distinct(wikientity) %>% pull(wikientity)
#
#
#
#
# fasttext_emb_test <- fastrtext::get_word_vectors(model, words = word)
# fasttext_emb_test <- fasttext_emb_test %>% as.VectorSpaceModel()
# rownames(fasttext_emb_test) <- word
#
# look_up <- function(){
# ent <- wordVectors::closest_to(entity_candidate_descr_embs, fasttext_emb_test) %>% rename(wikientity=word) %>% inner_join.(entity_overlap %>% distinct(wikientity, words)) %>% arrange(desc(`similarity to fasttext_emb_test`)) %>% as.data.frame() %>% pull(wikientity)
# ent[1]}
# safe_lookup <- possibly(look_up, NA)
#
#
# disambiguated_entity <- safe_lookup()
# rm(model, fasttext_emb_test, entity_candidate_descr_embs, test_entities)
# gc()
# disambiguated_entity <- data.table(disambiguated_entity=disambiguated_entity, words=word)
# disambiguated_entity
#
# }
#
# #options(future.fork.enable = TRUE)
#
# #library(furrr)
#
# #plan(multicore, workers=12)
#
# #results <- future_map_dfr(.x = overlapping_words, .f = disambiguate, .progress = T)
#
# results <- pbmcapply::pbmcmapply(X = overlapping_words, FUN = disambiguate, mc.cores = 15L) # works cleaner with forking
#
# results2 <- results %>% t() %>% as.data.frame()
results2 <- fread("/run/media/knut/HD/knut/Desktop/MLearningAlgoTests/data/polar/polar_quantized_fasttext_entities.csv")This should be almost it. Only gotta grab the knowledge graph embedddings.
results2 <- results2 %>% rename(wikientity=disambiguated_entity) %>% inner_join.(entities %>% distinct.(wikientity, rowid)) %>% inner_join.(polar_embeddings, by=c("rowid"="rowid"))You can find the embeddings (130k records) on Huggingface:
https://huggingface.co/datasets/KnutJaegersberg/interpretable_word_embeddings